New computational tool provides clues on how to target cancers with KMT2D mutations

Cancer is a genetic disease – it occurs when mutations or other genetic alterations take away cells’ normal safeguards and allow them to grow and divide unchecked.
In recent years, researchers have found that there are two main categories of genes that, when mutated, can lead to cancer: oncogenes and tumour suppressor genes. Oncogenes are genes that are inappropriately activated by mutations and whose supercharged activity can cause cancer. In contrast, tumour suppressor genes, as their name suggests, are safeguard genes that normally control functions that promote cancer development, such as unlimited growth. Inactivating mutations in these genes, which dial down or entirely eliminate the function of the genes, remove the restraints present in normal cells and can allow cancers to grow.
The difference between these two types of genes has important implications for treatment: in general, it’s easier to design a drug to block activated oncogenes than it is to design one that restores inactivated tumour suppressor gene functions. Consequently, many of the therapies we have available today inhibit the proteins encode by mutated oncogenes. Cancers driven by the loss or mutation of tumour suppressor genes can therefore be more difficult to treat effectively.
One way to target tumour suppressor gene mutations lies in a concept called “synthetic lethality”. Two genes are said to be “synthetic lethal” if the inactivation of either gene does not affect a cell’s ability to survive, but the inactivation of both genes in the same cell causes the cell to die. In the clinic, this concept was first exploited with a class of drugs called PARP inhibitors for patients whose cancers lack the BRCA1 or BRCA2 genes. PARP inhibitors were first approved by the US Food and Drug Administration (FDA) in 2015 for patients with advanced ovarian cancer and inherited BRCA1/2 mutations. Since then, more PARP inhibitors have been developed and approved for different indications.
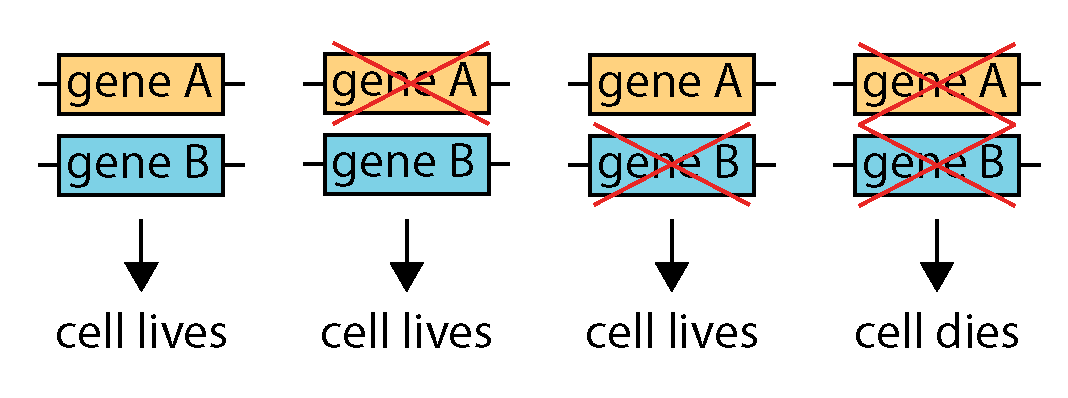
Diagram illustrating synthetic lethality.
Despite the general success of PARP inhibitors, identifying more synthetic lethal pairs that may lead to the development of new therapies is a challenging and expensive laboratory process. A new study published in Genome Medicine by members of the Marathon of Hope Cancer Centres Network proposes a new approach to identify synthetic lethal pairs using computational data (also known as an in silico analysis), instead of physical data.
The team, led by Dr. Marco Marra OC, OBC, PhD, FRS(C), FCAHS, and Terry Fox Leader in Cancer Genome Science at the University of British Columbia and Canada’s Michael Smith Genome Sciences Centre, used a tumour suppressor gene called KMT2D, which is frequently mutated in several cancer types, to demonstrate their approach. They used a tool recently developed by lead author Dr. Yuka Takemon (UBC) and Dr. Marra, called GRETTA, and leveraged a large publicly available dataset called the Cancer Dependency Map, or DepMap, to identify synthetic lethal interactors of KMT2D.
“This approach allows us to use data that have been made available to the research community and makes searching for synthetic lethal pairs more accessible and feasible at the level of individual labs,” says Dr. Takemon. “It helps us identify promising leads and dedicate more resources to validation and follow-up studies, increasing the likelihood that our findings will translate into a clinical benefit.”
Through their analyses, the team identified four genes that appear to be synthetic lethal interactors of KMT2D and that are targets of either existing or in-development drugs, providing a rationale for testing these therapeutics in cancers that lack KMT2D. In fact, inhibitors for one of these genes, WRN, are currently in phase I clinical trials. Results from this study could thus potentially help identify patients who are most likely to respond to these new drugs.
The team also found that, in a specific subtype of colorectal cancer, loss of KMT2D was associated with markers suggesting these cancers may respond well to immunotherapy. Although immunotherapy can be successful in some patients, researchers and clinicians don’t always know ahead of time whether or not a patient will respond. Identifying predictive markers, such as (potentially) KMT2D loss, can thus help stratify patients and provide them with the treatment from which they are most likely to benefit, reducing unnecessary treatments, associated toxicity, and costs.
“In this study, we introduce a framework for revealing potential vulnerabilities of tumour cells harbouring loss-of-function mutations in tumour suppressor genes,” says Dr. Marra. “Using KMT2D as an example, we demonstrate that our bioinformatics approach can be used to uncover potential novel biomarkers and treatment options. We believe this opens new opportunities to use genomic data to identify potential therapeutic avenues for difficult-to-treat cancers.”
Some of the sequencing data used in this study (from the Personalized OncoGenomics program) were partially funded by the Marathon of Hope Cancer Centres Network. These samples are now part of the MOHCCN Gold Cohort, which means that in the future, other researchers may use these data to further their precision medicine studies.
Learn more:
Click here to learn more about the Personalized OncoGenomics (POG) program.
Click here to learn more about Dr. Marra’s research, and here to learn more about Dr. Takemon and their work.
Full Citation:
Yuka Takemon, Erin D. Pleasance, Alessia Gagliardi, Christopher S. Hughes, Veronika Csizmok, Kathleen Wee, Diane L. Trinh, Ryan D. Huff, Andrew J. Mungall, Richard A. Moore, Eric Chuah, Karen L. Mungall, Eleanor Lewis, Jessica Nelson, Howard J. Lim, Daniel J. Renouf, Steven J.M. Jones, Janessa Laskin and Marco A. Marra. Mapping in silico genetic networks of the KMT2D tumour suppressor gene to uncover novel functional associations and cancer cell vulnerabilities. Genome Medicine (2024) 16:136 doi: 10.1186/s13073-024-01401-9
“We believe this bioinformatics approach opens new opportunities to use genomic data to identify potential therapeutic avenues for difficult-to-treat cancers.”
Related Team Members
-
Marco
ResearcherMOHCCN Network CouncilWorking Group ChairConsortium LeaderWorking Group Member
Marra -
Yuka
Working Group Member
Takemon
Related Project
-
Regional Consortia
BC Cancer Consortium
- British Columbia
The overall vision of the BC Cancer Consortium is to further strengthen our province-wide research capabilities and partnerships, to improve our ability to support innovation in clinical care, and fir...Read more
-
Regional Consortia
BC Cancer Consortium
- British Columbia
The overall vision of the BC Cancer Consortium is to further strengthen our province-wide research capabilities and partnerships, to improve our ability to support innovation in clinical care, and fir...Read more
-
Regional Consortia
BC Cancer Consortium
- British Columbia
The overall vision of the BC Cancer Consortium is to further strengthen our province-wide research capabilities and partnerships, to improve our ability to support innovation in clinical care, and fir...Read more
-
Regional Consortia
BC Cancer Consortium
- British Columbia
The overall vision of the BC Cancer Consortium is to further strengthen our province-wide research capabilities and partnerships, to improve our ability to support innovation in clinical care, and fir...Read more
-
Regional Consortia
BC Cancer Consortium
- British Columbia
The overall vision of the BC Cancer Consortium is to further strengthen our province-wide research capabilities and partnerships, to improve our ability to support innovation in clinical care, and fir...Read more
Related News
-

Atlantic Cancer Consortium leadership and members share updates with the Network
The presentation was part of MOHCCN Seminar Series, which features virtual talks from MOHCCN research partner investigators and trainees, providing a platform for MOHCCN members to share their ongoing... -

New study highlights long-read sequencing applications in precision oncology
A team of researchers that includes Marathon of Hope Cancer Centres Network members demonstrate applications for long-read sequencing in precision oncology in a new study published in Cell Genomics. -

Prairie Cancer Consortium leadership and members share updates with the Network
The presentation was part of MOHCCN Seminar Series, which features virtual talks from MOHCCN research partner investigators and trainees, providing a platform for MOHCCN members to share their ongoing... -

New study explores patient preferences and concerns to improve data sharing for precision oncology across Canada
A team of health researchers that includes MOHCCN members conducted focus groups with individuals with lived and living experience with cancer to understand their values, expectations, and concerns fo...


